 Welcome to my website! Welcome to my website!
I am a post-doctoral Research Fellow at the Department of Systems Biology in Harvard Medical School working with Prof Jeremy Gunawardena.
I did my PhD in Neuroscience at the University of Cambridge with Prof Sabine Bahn. I took a functional genomics approach to understand schizophrenia using different 'omics' platforms to identify potential disease signatures in pre-frontal cortex area of the brain. It was during my PhD that I realized the importance of looking at the dynamics of protein interactions to reveal more about cellular mechanisms than looking at snap shots of protein interactions from 'omics' data.
In my post-doctoral work with Prof Gunawardena, I looked at protein-protein interactions with model protein biochemical systems with simple modification and de-modification cycles such as phosphorylation and de-phosphorylation. These simple systems have been analyzed in detail in the past by eminent scientists like Goldbeter and Koshland. However when the modification and demodification happens on more than one site, what we define as 'multisite' modification, the dynamics of protein interactions becomes very complicated and almost difficult to predict.
Proteins have been traditionally conceived as 'on/off' switches, however multisite modifications creates intermediate states of proteins with a total potential of 2^n states, where n is the number of modification sites. Hence a protein population is actually a relative distribution of a protein among all these states. Different relative distribution of states of a protein might induce different cell fate decisions. My specific interest is to understand the regulation of proteins by multisite modifications using Mass Spectrometry, NMR and Mathematical modelling and to unravel the biological role of distibutions of protein states.
My over all interest is to understand how cells achieve signaling specificity in spite of using (or re-using) a small number of proteins in multiple signal transduction pathways. More importantly, I am interested in what is called as ‘context specificity’ where the same proteins (cataloged to be involved in a particular signaling pathway) make different cell fate decisions depending on cell-type or differentiation or developmental or cell-cycle stage. Proteins that play a crucial role in the signal transduction cascades are actually multiply modified from receptors to transcription factors. It is possible that different distributions of states of a protein might determine the down stream responses giving rise to signaling specificity. I believe, by understanding regulation and role of multisite post translational modifications we would be able to understand signaling specificity. |
Research
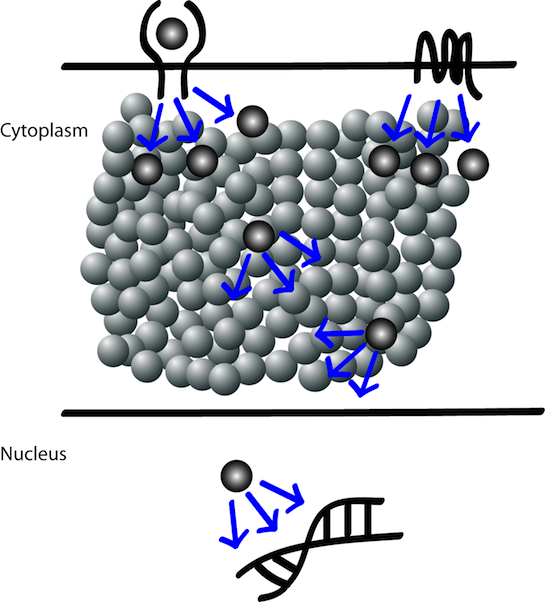
Cell fate decisions are being made at every stage and state of signal transduction events. From receptors at the surface to signaling proteins in the cytoplasm and transcriptional factors in the nucleus every component of a signal transduction cascade are involved in cell fate decisions (more...). |
|
Background
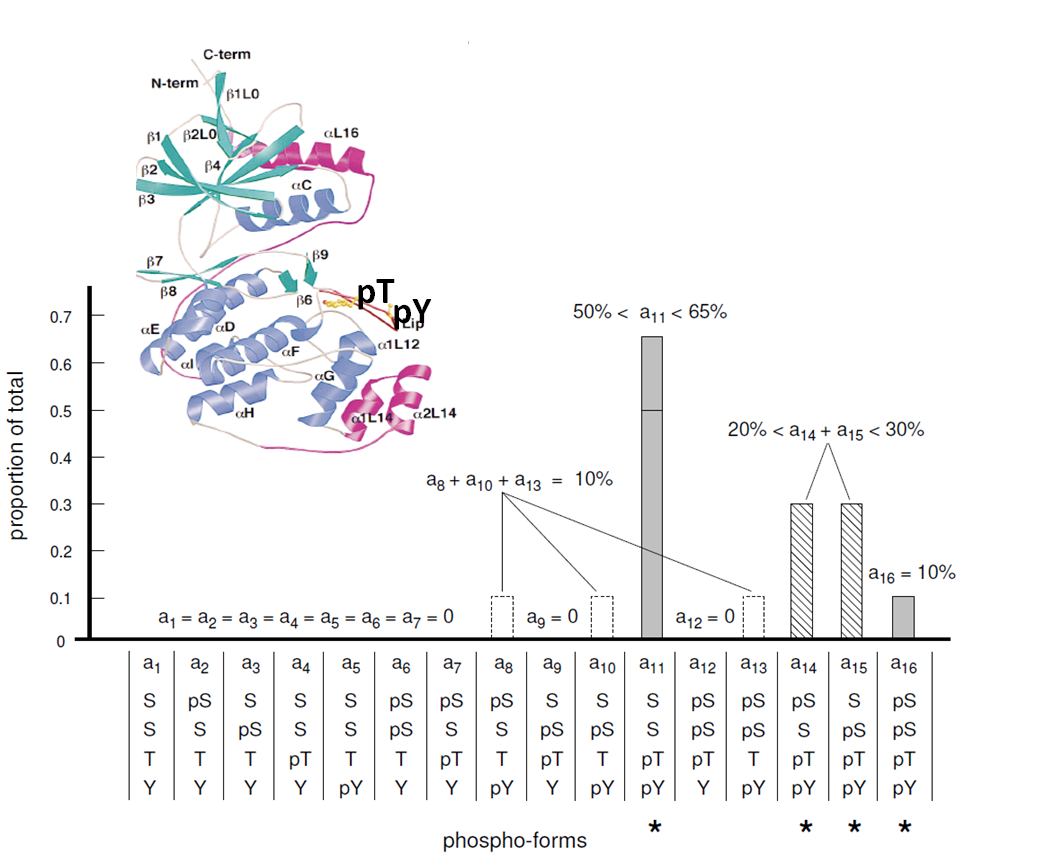
In my post doctoral work, my collegues and I were able to show that it was in principal possible to unravel the distribution of all the 16 possible phospho-form states of a signaling protein (Erk2) with 4 phosphorylational sites (of which 2 were novel findings) using peptide-based, protein-based Mass Spectrometry and NMR (more...). |
Biography
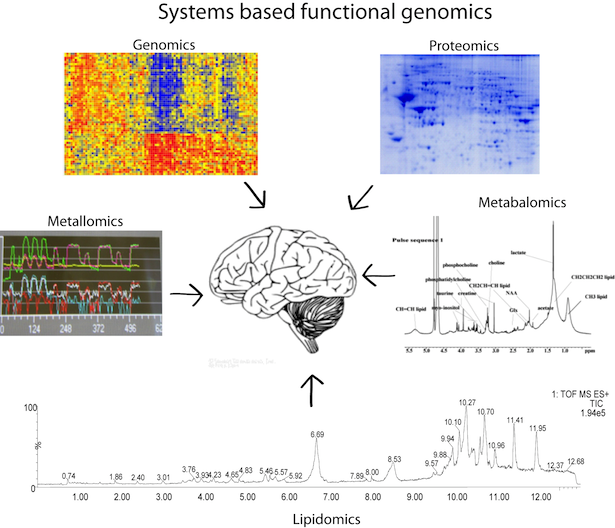
In my doctoral work, I used a sytems-based 'multi omic' strategy to unravel schizophrenia disease signature in the pre-frontal cortex of human brain. I used genomic (gene arrays), proteomic (2D DIGE and protein mass spectrometry), metabalomic (NMR), metallomic (Laser ablation Inductively-coupled plasma mass spectrometry and lipidomic (lipid mass spectrometry) platforms (more..._). |
 Welcome to my website!
Welcome to my website!

